Tutorial
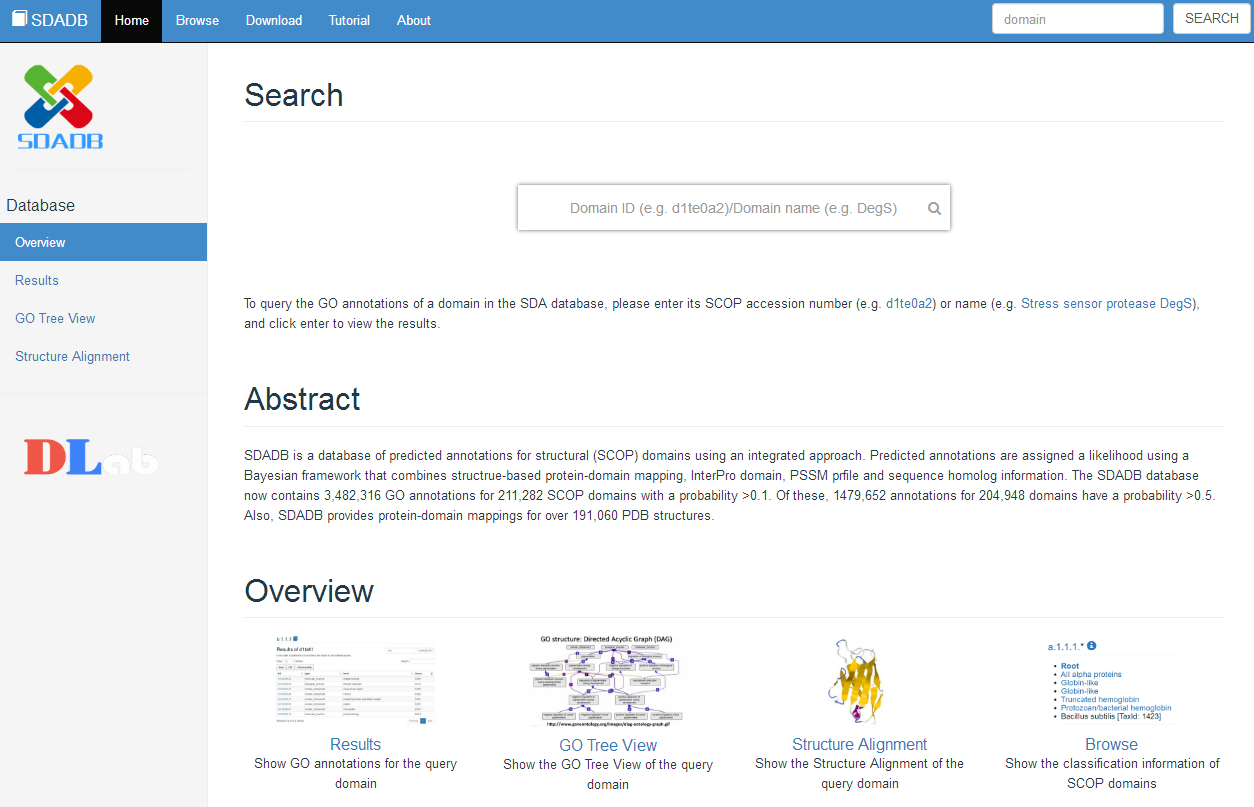
Overview view
The index page is mainly show the overview of our SDADB. We provide a query input in this page to search the domain name, and you can also search in the right of the top menu. Both of them has the same function, and if you are interested in some domain but not sure the accurate?name of the domain, you can also search the protein name or the classification of the domain. Beside the search function, we also show the abstract and the main function of our SDADB below the query input.
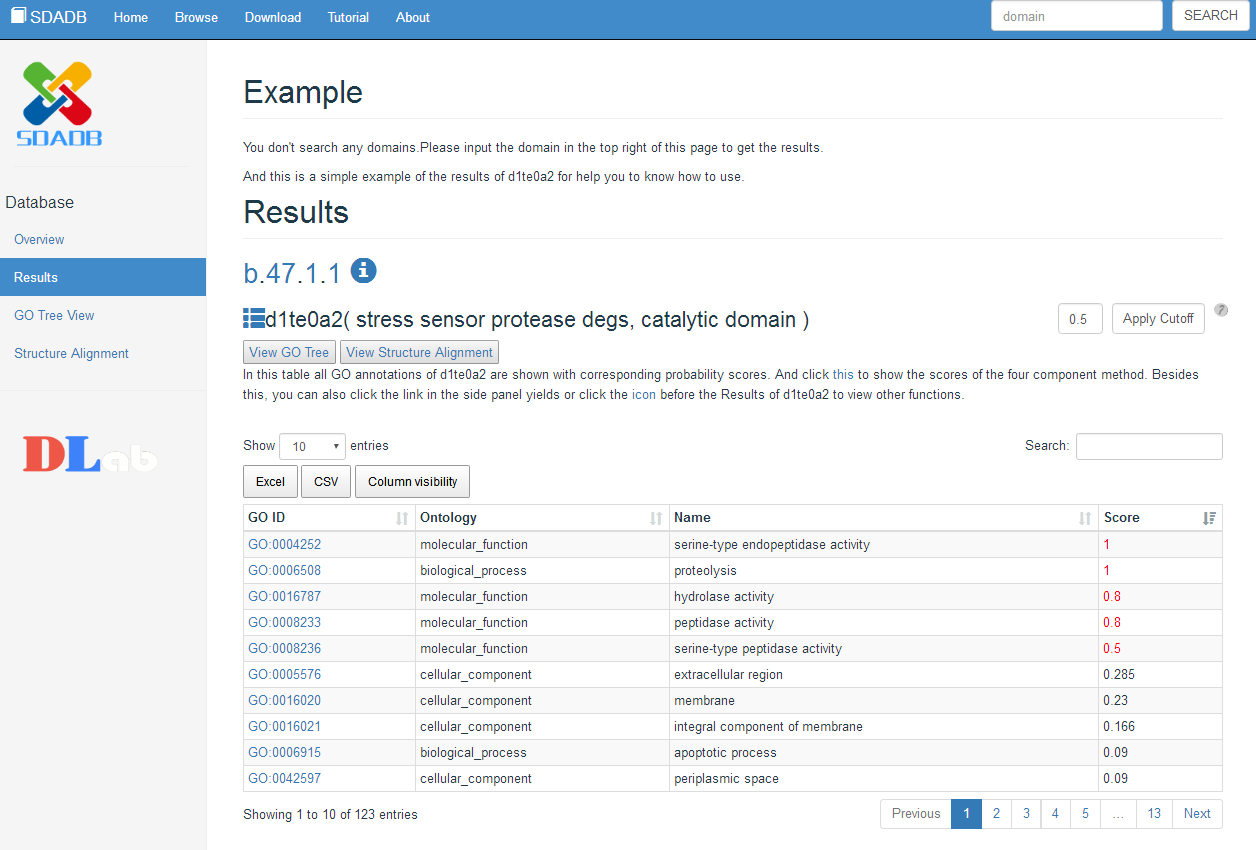
Results view
After you search the name of the domain, we provide the Results page. And in this page, we mainly show the predicted results for the search domain. All of the annotation information is show in the table. Including the GO ID, GO type, GO name and the predicted score. The table also provide the search function to get the target GO and the download function to get the results in excel file or csv file. And to show the difference of the high score result,all the score that over the cutoff is in red color, the default cutoff is 0.5, and you can change the cutoff according to your requirement by input the cutoff in the above the search input.
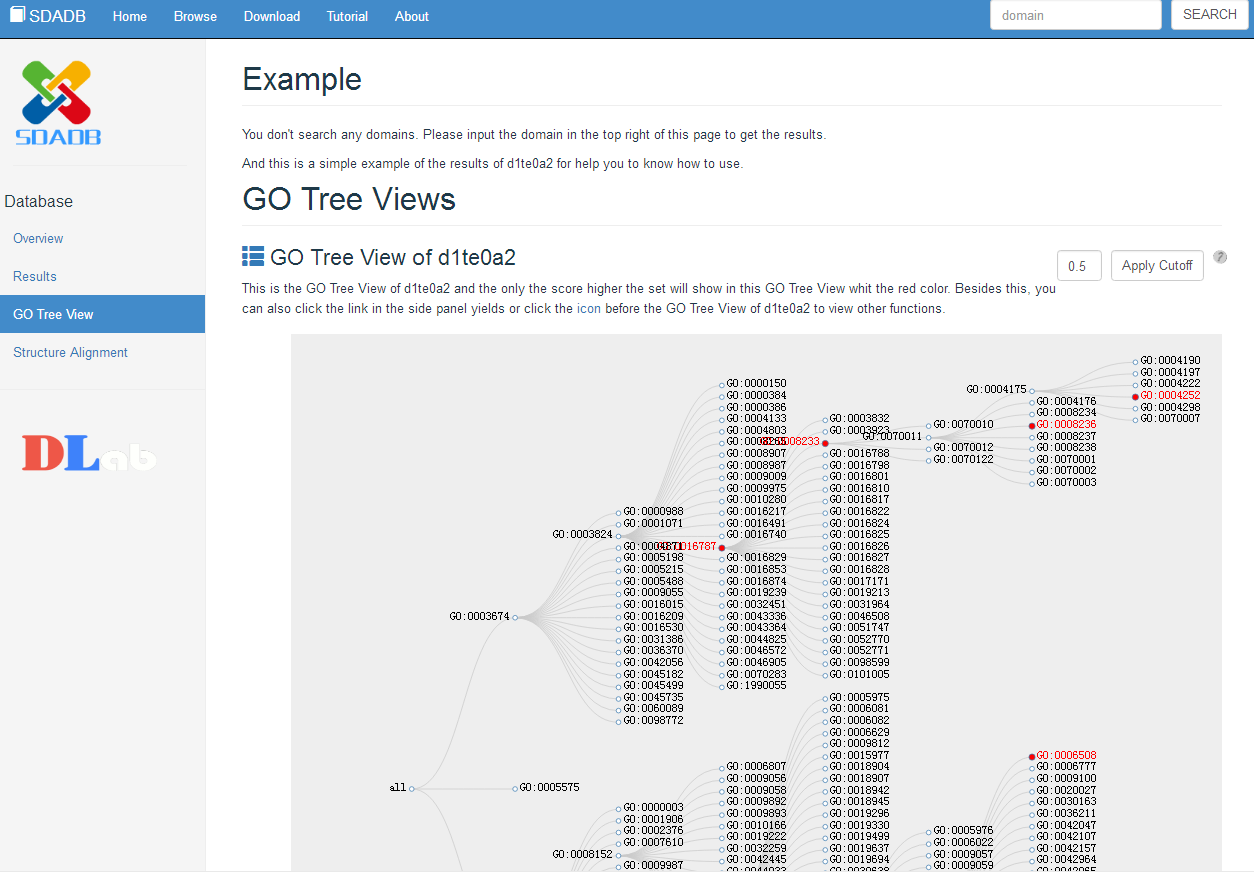
GO Tree View view
We also provide the GO tree view to show the relationship of the predicted GO terms. You can see the GO tree by clicking the GO trees in the side panel yields. To show more accurate and more clearer relationship, only the GO that score over the cutoff will show in this GO tree with red color.
Alignment View
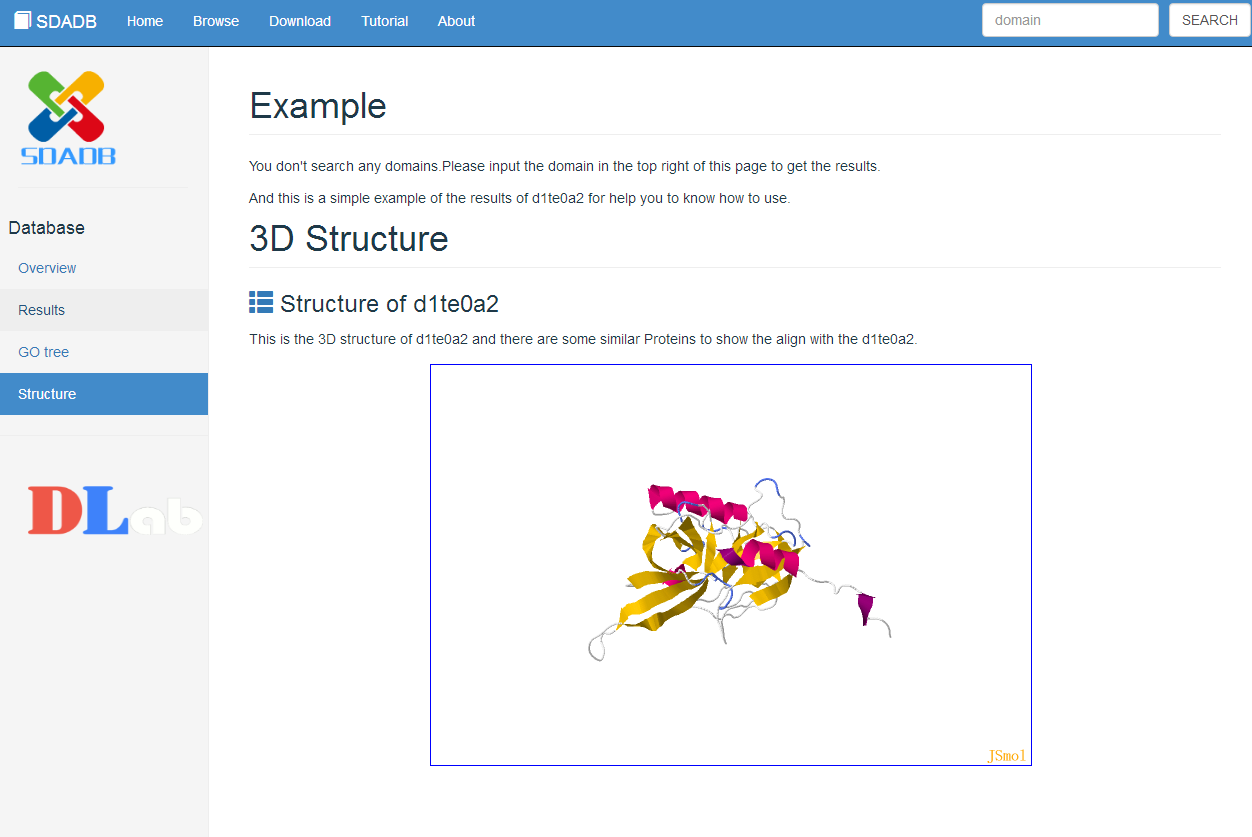
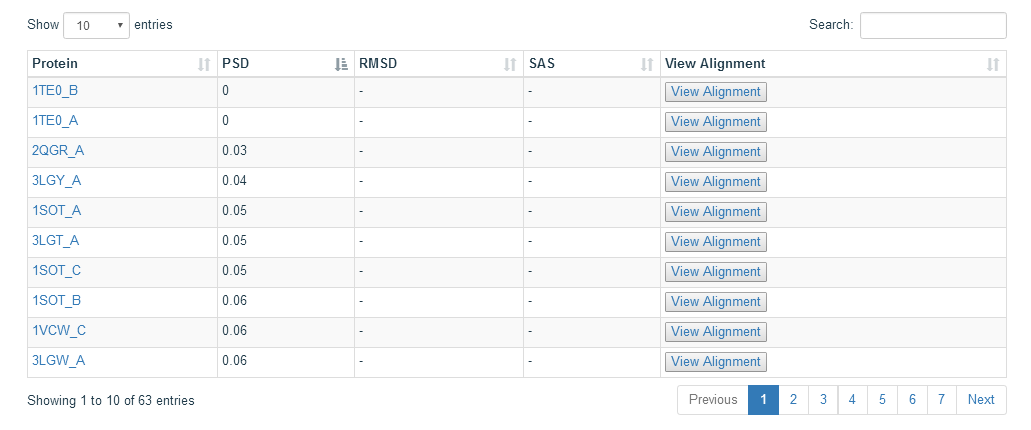
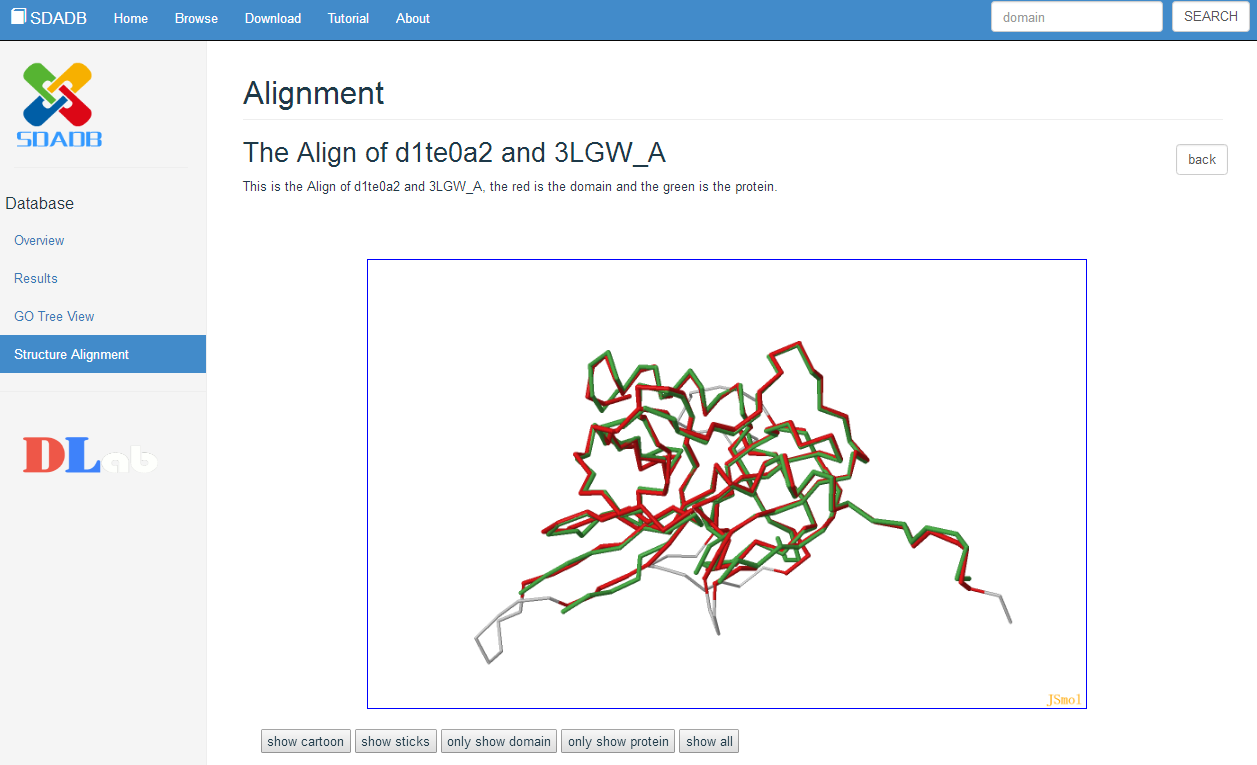
To show more information, we also provide the 3D structure view and the 3D alignment view of the domain. The proteins mapping to the search domain are show in the table below the 3D structure view. The structural neighbor proteins are obtained by using large-scale all-against-all structure alignments to obtain mappings between PDB structures and SCOP domains. Only the proteins with PSD score less than 0.1 are shown in the table. The PSD score means the score more close to zero, the proteins more similar to the search domain.
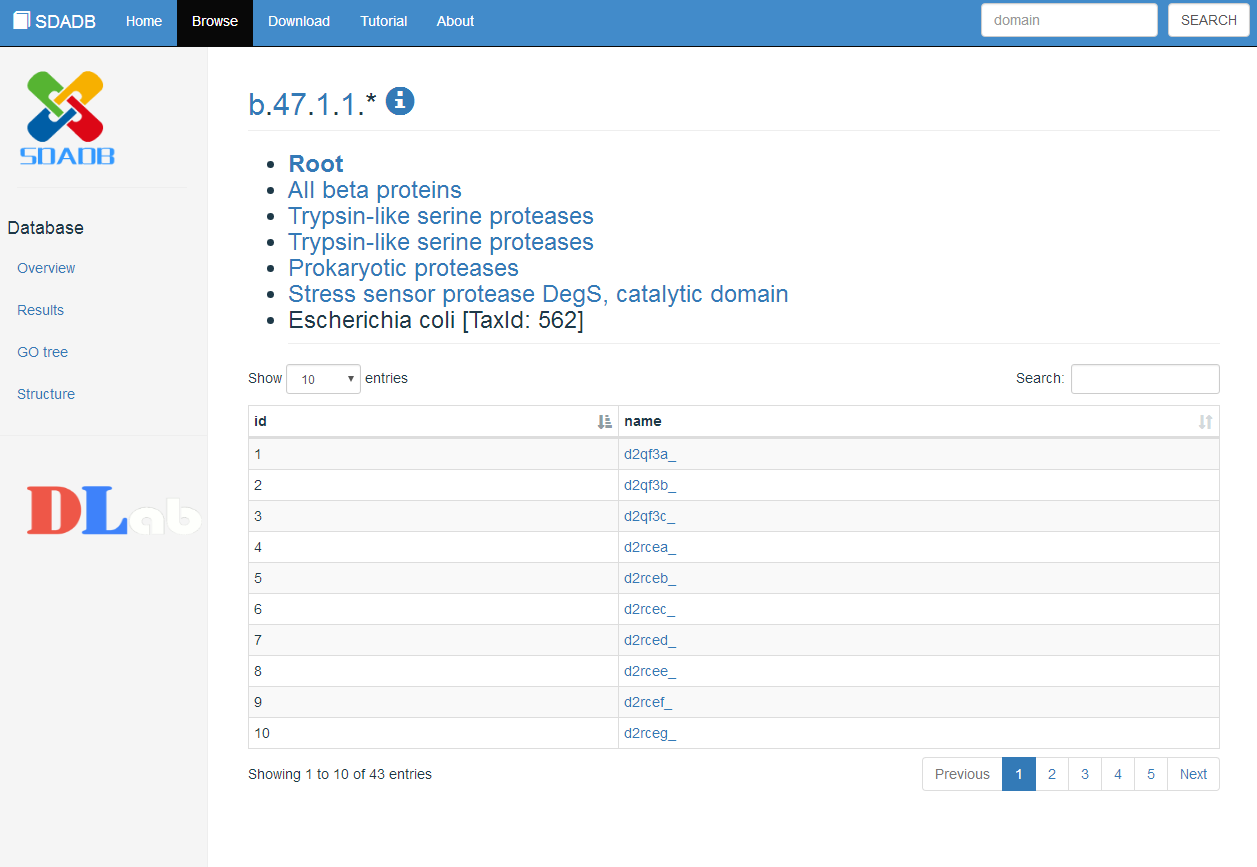
Browse view
We also provide the browse function to show the classification of those domains. The classification information of those domains is obtained from the SCOPe databases including Class, Fold, Superfamily, Family, Protein and Species.