About
SDADB: a functional annotation database of structural domains in proteins
Currently, there are more than 244,326 distinct structural domains as represented by the unique folds in the SCOP database. In order to annotate the functions of those structural domains, we present a database named Structural Domains Annotation Database. In the SDADB database, we gather heterogeneous information sources and integrated by a Bayesian network to obtain better result. The SDADB database not only present the structural domains and their annotations, but also include 3D structure alignment visualization, GO hierarchical tree view, search, browse and download options.
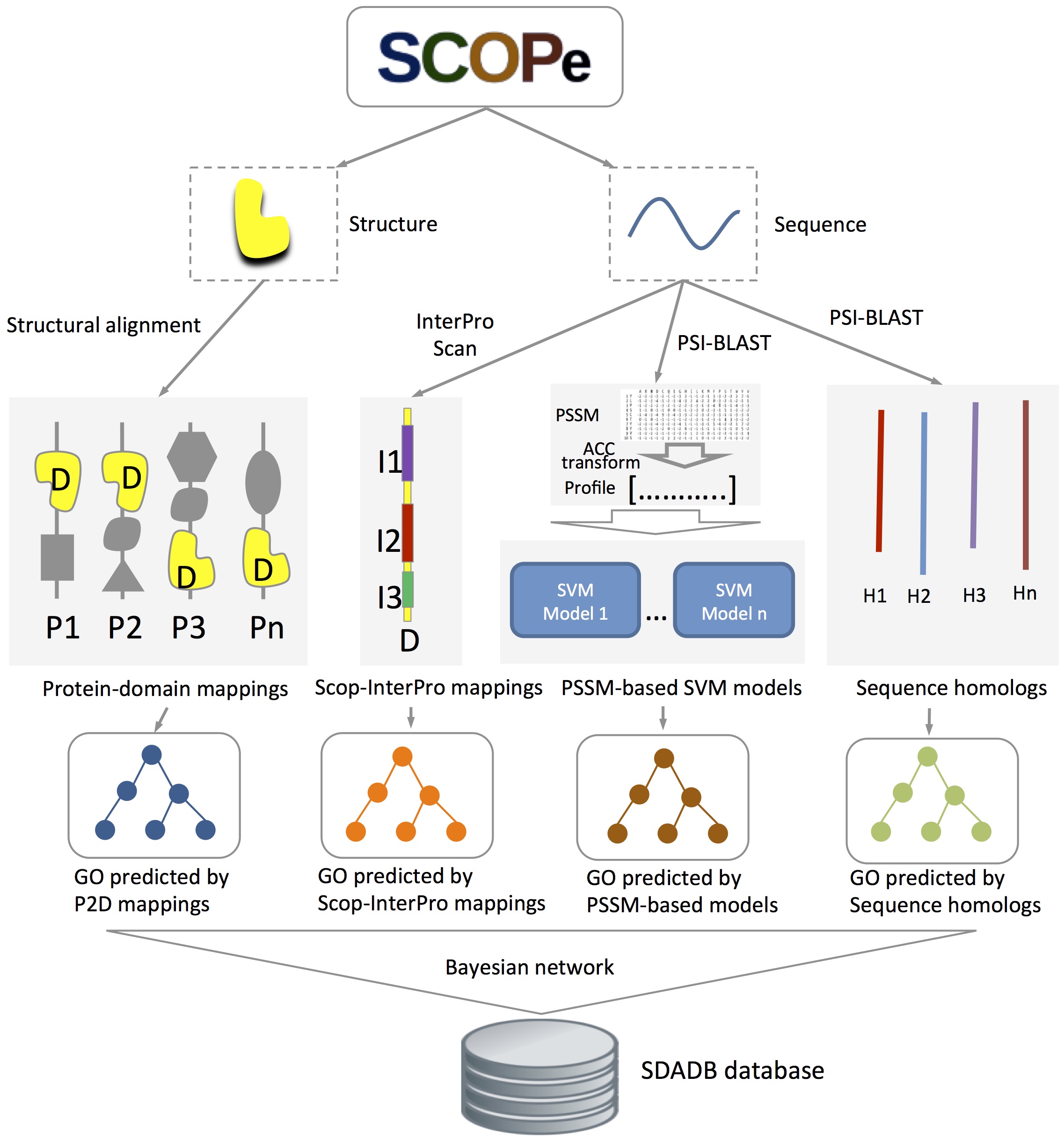
Figure 1 shows the Framework of the SDA method which we use in this databases. For a query domain, annotations are predicted with four component methods: 1) structure-based method: protein-SCOP domain mapping information obtained from all-against-all structure alignment is used to calculate the probability that a domain annotated by a specific function; 2) InterPro-based method: InterPro domains of a SCOP domain sequence are extracted using the InterProScan tool, annotations of these InterPro domains from InterPro2GO database are transferred to the SCOP domain; 3) position-specific scoring matrices based method: SVM models for GO function annotation are trained with fixed length of vectors, which are generated from SCOP domain's PSSMs with auto-cross covariance transformation; and 4) sequence homolog-based method: functions of SCOP domains are transferred from their sequence homologs'GO annotations in UniProt-GOA. Finally, a Bayesian network is used to combine the outputs of the four methods.
Authors
- Cheng Zeng
- Lei Deng
Contact
If you have any questions or suggestions about the SDADB database, do not hesitate to contact us.
Options
References
- [1] An Integrated Framework for Functional Annotation of Protein Structural Domains
-
An integrated framework for functional annotation of protein structural domains.
IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB).
2015.
doi: 10.1109/TCBB.2015.2389213.
BibTeX
@article{deng2015integrated, title={An integrated framework for functional annotation of protein structural domains}, author={Deng, Lei and Chen, Zhigang}, journal={IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB)}, volume={12}, number={4}, pages={902--913}, year={2015}, publisher={IEEE Computer Society Press} }
